L’Acide Case: Cleanup Recommendation Report
This page includes the Cleanup Recommendation Attachment to accompany handouts for the L'Acide case discussion. It includes necessary background information for the case.
Download this report in pdf format.
Background information for the City of L'Acide Remediation Case
DO NOT CITE OR QUOTE!
Technical Recommendation
The remediation will consist of three stages:
- Installation of a slurry wall to contain the plume.
- Injection of surfactants to desorb the recalcitrant contaminants from soil.
- Injection of bacterial cultures to degrade the compounds in situ.
Based on the soil type and other local conditions, Benebaction recommends that microbial growth and metabolism, as well as that of larger organisms, will be the most efficient choice for degradation. In fact, we recommend a specific culture of Pseudomonas etemup, which has been shown in laboratory studies to degrade nitrogenous compounds rapidly, so we expect it to be ideal here. The strain that has to be used (Booboom A) is genetically modified (plasmid insertion) to use N-compounds as its food source.
Benebaction also recommends installing a white rot fungus (Phanerochaete chryosporium) bioreactor for all extracted materials (mainly soils) on site. The P. chryosporium will also be genetically enhanced, as will the sage grass to be planted around the site (to be weed resistant).
Background
Benebaction, Inc. seeks “green solutions”, and since microbial and other biological populations are essential parts of natural systems, we have gained an excellent reputation at using biotechnologies to emulate these processes in engineered systems. Traditionally, indigenous soil bacteria have been encouraged to use some rather exotic chemical compounds as food, especially when it is the sole source of carbon and electrons (i.e. acclimation).
Some compounds are recalcitrant, refusing to be the carbon and energy sources for most microbes. Sometimes, the solution lies in finding a different type of organism. The keen observations of environmental engineers and biologists led to the logical use of fungi, since these organisms are quite proficient at degrading the otherwise recalcitrant natural polymers, especially lignin in wood. Bacteria have a much harder time degrading these substances, but bioscientists reckoned that different orders of organisms could be put to use. One observation was that thousands of strains and species of white rot fungi (including Phanerochaete chryosporium) have been shown to degrade lignin, cellulose, and hemicellulose. Taking this into account, P. chryosporium has been put to work in increasingly larger reactors (from lab benches to full scale ex situ operations). In addition to aromatic hydrocarbons and other organic compounds, fungi have also been used to extract and detoxify metallic contaminants.
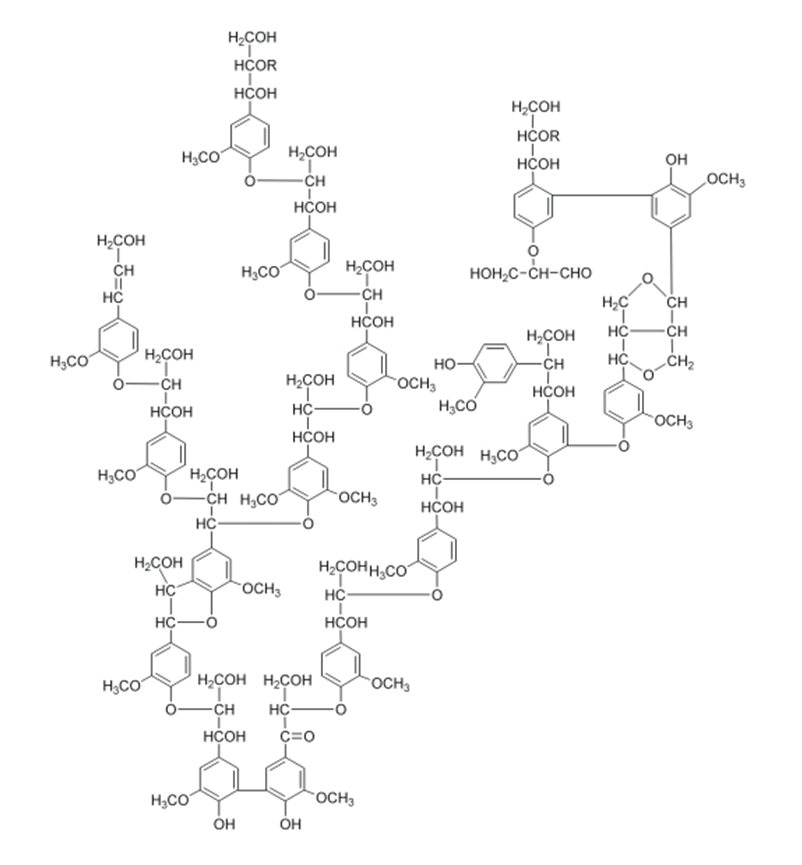
Both cellulose and lignin are polymers, which are large organic molecules comprised of repeated subunits (i.e. monomers). However, lignin is naturally recalcitrant, owing to its covalent bonds to hemicellulose and cross-linking to polysaccharides which hold woody tissue together and provide the rigidity of wood compared to non-lignin containing plants (See Fig. 1). Lignin fills the spaces in a woody plant’s cell wall between cellulose and two other compounds, hemicellulose and pectin. The monomers that comprise lignin polymers can vary, depending on the sugars from which they are derived. In fact, lignin polymers contain so many random couplings that the exact chemical structure is seldom known.[1]
Lignin is not a source of energy for white rot fungi, so their biodegradation of toxic pollutants follows the prototypical cometabolic pathway, i.e. they need other substrates (e.g. cellulose). This need, however, makes fungi particularly attractive to green engineering solutions (e.g. composting). That is to say, silage and other vegetative waste material from agricultural operations can be degraded, and in the process, enhance degradation rates of xenobiotic contaminants in polluted material, either in situ or ex situ at polluted sites. All plants contain cellulose, but woody plants also contain lignin.
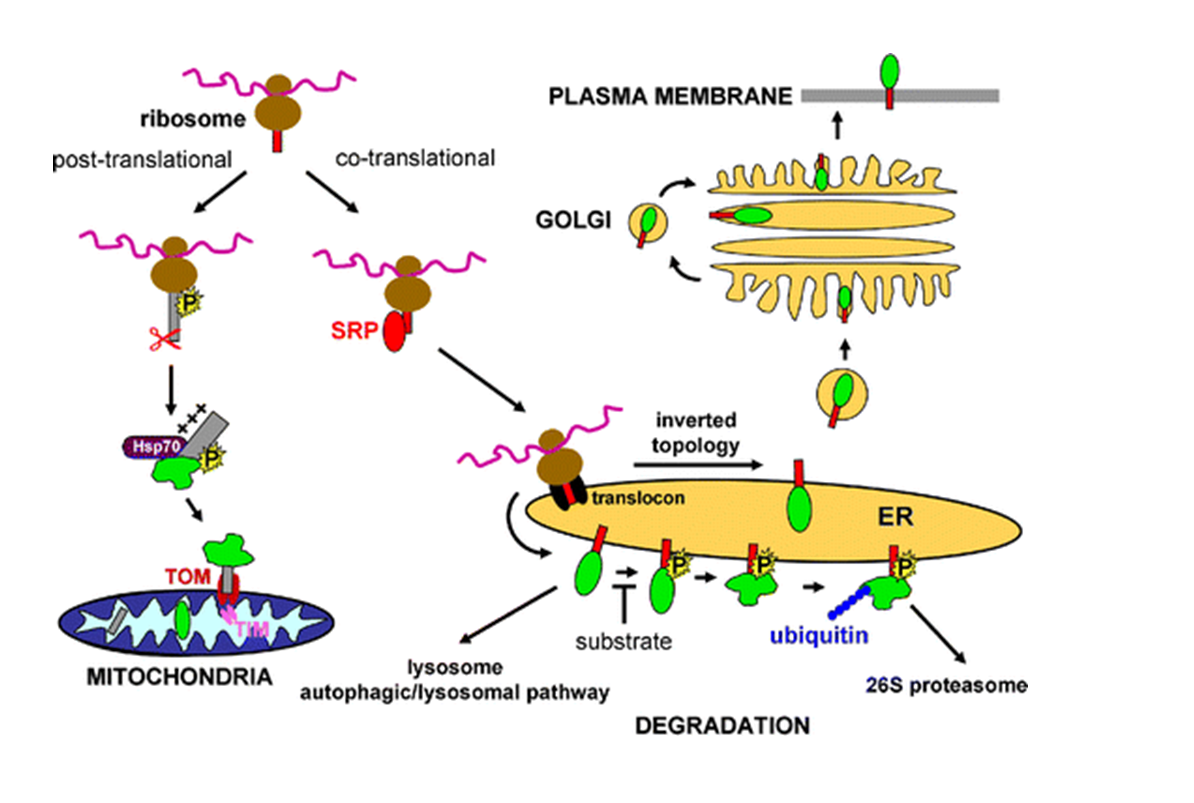
Fig. 1. An example of possible configurations of a lignin polymer. Source: Institute of Biotechnology and Drug Research, “Environmental Biotechnology and Enzymes,” Kaiserslautern, Germany. Adapted from: E. Adler, (1977). Lignin chemistry-Past, Present and Future, Wood Science Technology. 11: 169-218. The systematic perspective takes into account that white rot and other fungi branch and form filaments, which indicates that their growth is especially conducive to soil treatment. The principle mechanism for white rot fungus degradation is enzymatic, especially the fortuitous lignin degradation system of enzymes. Extracellular lignin modifying enzymes (LMEs) are not very substrate specific, which enables them to mineralize numerous, otherwise recalcitrant contaminants, particularly those with chemical structures similar to lignin.[2] The predominant LMEs are lignin peroxidase,[3] manganese (Mn)-dependent peroxidase,[4] and laccase.[5] The extracellular enzymatic mechanism allows for contact with numerous organic contaminants with low aqueous solubility (e.g. chlorinated and multiple ring aromatic compounds) not allowed when restricted to intracellular processes. For example, catalysis by cytochrome P450 would first require that the contaminant compound be transferred across the microbe’s membrane, followed by contact with the cytochrome enzymes (See Fig. 2). These are predominantly localized in the endoplasmic reticulum (ER) but can also be found in other subcellular compartments, such as mitochondria, plasma membrane, and lysosomes.[6]
Fig. 2. Intracellular targeting, transport, and degradation of microsomal cytochrome P450. Normally, cytochromes P450 are targeted to the endoplasmic reticulum (ER) via a signal recognition particle (SRP) process. A very small quantity of the enzymes may be inserted inversely and transported via the Golgi apparatus to the plasma membrane. Cytochromes P450 are degraded via the lysosomal pathway or after phosphorylation (P) or covalent modification by the proteasomal pathway. The presence of a substrate can prevent phophosporylation and subsequent proteosomal degradation. Modification of the signal sequence by phosphorylation or proteolytic processing (shown as scissors) prevents efficient binding of the SRP that leads to translation of the entire protein in the cytoplasm and after interaction with chaperones (Hsp70) to mitochondrial targeting and import. TOM translocase of the outer membrane, TIM translocase of the inner membrane Cytochromes P450 can be degraded via the lysosomal pathway or after phosphorylation (P) or covalent modification by the proteasomal pathway. The presence of a substrate may prevent phophosporylation and subsequent proteosomal degradation. Source: E.P.A. Neve and M. Ingelman-Sundberg (2008). Intracellular transport and localization of microsomal cytochrome P450. Analytical & Bioanalytical Chemistry. 392 (6): 1075-1084.
Cometabolism is an excellent illustration a biotechnological systems approach. On its own, each species in a successful bioremediation project may not solve the problem. However, the synergies among species and strains of microbes can lead to successes not possible from reductionism. The metabolism of a microbe that does not need our targeted contaminant as a carbon source will nonetheless lead indirectly to detoxification due to degradation that is catalyzed by an enzyme that is fortuitously produced by the organisms for other purposes. The microbe does not directly benefit from the degradation of the compound.[7] In fact, the biotransformation of the compound could actually be harmful or can inhibit growth and metabolism a microbe. This process would make little sense from a reductionist viewpoint, but is perfectly reasonable from the systematic perspective.
Thus, we can add a second or additional carbon sources (e.g. unsubstituted alkanes) that stimulate biodegradation of the targeted pollutant. For example, halogenated compounds that are inherently more recalcitrant than their non-halogenated counterparts can be degraded by stimulating soil bacteria with the addition of O2 and CH4 to the subsoil strata.
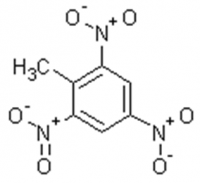
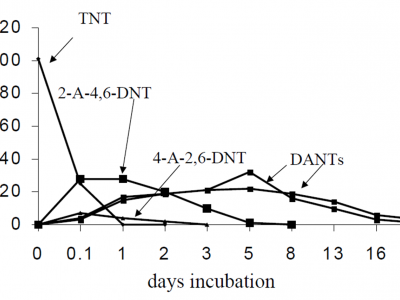
Our biggest challenge is the explosive trinitrotoluene (TNT). Obviously, the compound degrades extremely fast when ignited, i.e. the explosion from the addition of oxygen in the presence of a heat source. This extremely rapid oxidation produces CO2, H2O and oxides of nitrogen. Scientists also observed that in the environment TNT can be reduced slowly. In laboratory studies, the majority of the TNT degrades to monoaminodinitrotoluene and diaminonitrotoluene isomers within a few days (See Fig. 3.).
Trinitrotoluene
The persistence of TNT in environmental systems is quite different. When released into the atmosphere it is degraded by predominantly direct photolysis, with an estimated atmospheric half-life ranging from 18.4 to 184 days.[8] These T1/2 estimates are based on the expected reactions with hydroxyl radicals in the atmosphere. The transformation of TNT in surface waters by microbial metabolism is much slower than photolysis. Under anaerobic and aerobic environments, the predicted biodegradation T1/2 of TNT in surface water ranges between 1 and 6 months.[9]
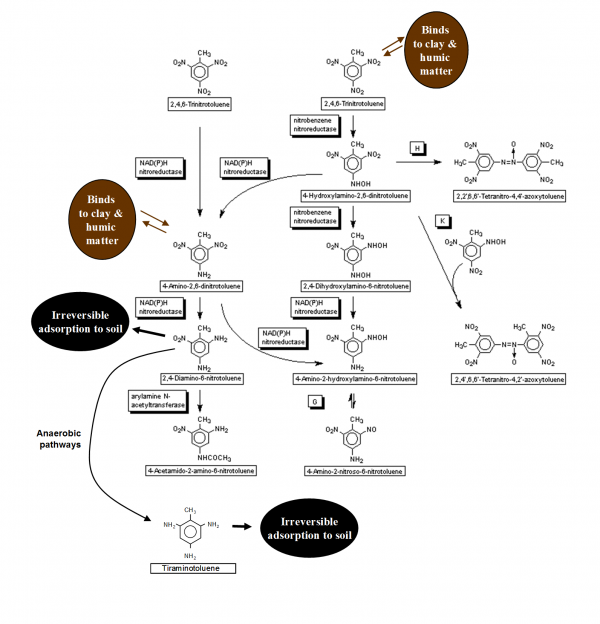
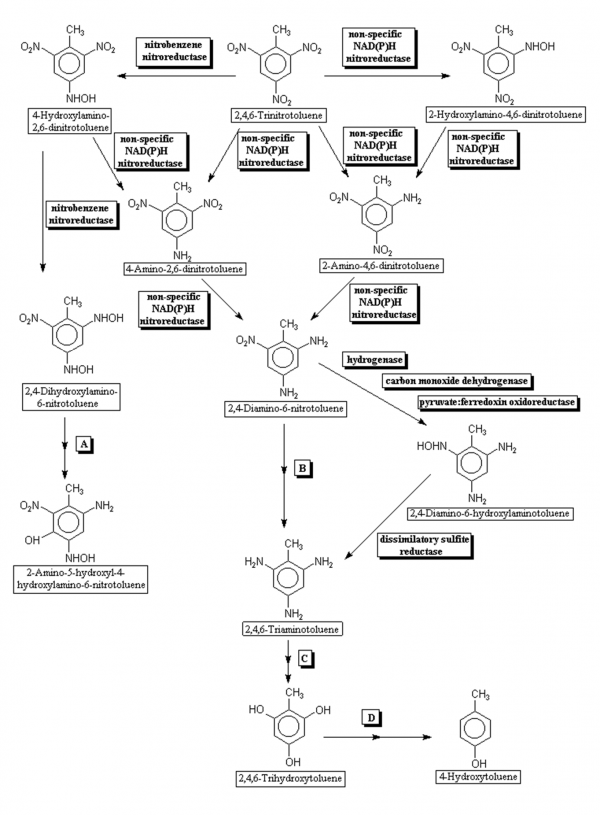
These rates do not represent environmental biodegradation, however, since environmental fate depends on more than mere abiotic chemistry. In addition to redox reactions, the TNT is sorbed to clay and organic molecules (e.g. humic acid) in the soil, with the intermediate degradates formed along the way. That is, TNT initially undergoes oxidation to form a variety of reduction products, culminating in the formation of triaminotoluene, which can be irreversibly adsorbed to soil’s clay and organic matter content (See Fig. 4). These chemical and physical mechanisms, i.e. redox and soil binding, call for a series of aerobic, then anaerobic degradation processes (See Fig. 5) to completely remove and degrade TNT when it is in soil. The reduction of one nitro (-NO2) group of TNT is quite fast by aerobes. Conversely, the reduction of 2-amino-4,6-dinitrotoluene requires a lower redox potential, and reduction of 2,4-diamino-6-nitrotoluene requires a very low redox potential (i.e. < -200mV), due to the electron-donating properties of the amino (-NH2) group’s decreasing the electron deficiency of the molecule.[10]
TNT is biodegraded aerobically by a number of organisms, including fungi (Phanerochaete chryosporium and Irpex lacteus), yeasts (Candida and Geotrichum spp.) and bacteria (e.g. Actinomycetes spp., Pseudomonas spp. and Alcaligenes 1-15), and anaerobically by bacteria (e.g. Methanococcus spp. B strain[11], Desufovibrio spp., Clostridium pasteurianum and Moorella thermoacetica[12]).
Incidentally, cometabolism appears to enhance TNT’s degradation rates (See Table 1). The cosubstrates’ presence may enhance the TNT degradation rates by serving as H2 donors. Complex substrates can be fermented by microorganisms in a bacterial consortium, thus generating H2 as one of the products, which in turn becomes available to the TNT degrading bacteria.[13]
|
Electron Donor |
Trinitrotoluene degradation |
|
None |
2.2 |
|
Acetate |
2.7 |
|
Ethanol |
4.2 |
|
Glucose |
6.3 |
Table 1. Enhancement of biodegradation of trinitrotoluene (TNT) by addition of cosubstrates. Source: P. Hwang, T. Chow and N.R. Adrian (1998). Transformation of TNT to triaminotoluene by mixed cultures incubated under methanogenic conditions. U.S. Army Corps of Engineers. USACERL Technical Report No. 98/116.
Fig. 3. Degradation of trinitrotoluene (TNT) biodegradation in serum bottles incubated under reduced (methanogenic) conditions. Note: 2-A-4,6-DNT = 2-amino-4,6-dinitrotoluene; 4-A-2,6-DNT = 4-amino-2,6-dinitrotoluene; and DANTs = diaminonitrotoluene isomers. Source: P. Hwang, T. Chow and N.R. Adrian (1998). Transformation of TNT to triaminotoluene by mixed cultures incubated under methanogenic conditions. U.S. Army Corps of Engineers. USACERL Technical Report No. 98/116.
Fig. 4. One set of aerobic (e.g. Pseudomonas savastanoi) biodegradation pathways of trinitrotoluene, including influence of soil binding. Anaerobic pathway details are shown in Fig. 12.22. Sources: A. Scragg (2004). Environmental Biotechnology. Second Edition. Oxford University Press. Oxford, United Kingdom; and S. McFarlan (2009). 2,4,6-Trinitrotoluene Pathway Map. University of Minnesota Biocatalysis/Biodegradation Database.
Fig. 5. Anaerobic biodegradation pathway for trinitrotoluene. Most reactions are catalyzed by non-specific NAD(P)H dependent nitroreductases. The last reduction steps to produce triaminotoluene occur only under anaerobic conditions catalyzed by enzymes in Desufovibrio spp., Clostridium pasteurianum, and Moorella thermoacetica. These nitro group reductions are also catalyzed by purified xenobiotic reductase enzyme. Source: S. McFarlan and G. Yao (2009). Anaerobic Trinitrotoluene Pathway Map. University of Minnesota Biocatalysis/Biodegradation Database.
There is little debate about the enhancement of indigenous bacteria, yeast or fungi, to break down these unwanted contaminants. Adding oxygen, nutrients and water to improve rates of biodegradation is good engineering practice.
We recognize that when indigenous bacteria are acclimated to use otherwise unattractive food sources, the bioengineer is changing the genetic makeup of the microbial population. One pivotal concern is that of containment. There are numerous dimensions to containment, but they either have to do with the biological agent or the environment, and usually both. If the agent does not elicit real or perceived hazards, containment would not be an issue. And, even if an agent is considered hazardous, if there is no means by which it will move away from a “safely” localized site, it is unlikely to be of much concern.
In this case, the toxicity of TNT warrants a well-designed bioremediation project as a necessary cleanup action. We recognize that some ecologists may see the introduction of any unknown species as potentially irreversible, and from a precautionary perspective, it should be avoided if another organism or method will do the job. In this instance, abiotic degradation (e.g. ex situ incineration) may be more attractive to some. Clearly, though, the less the difference in time and costs between native and genetically modified strains the greater the advantage of native over the modified biota. If there is no difference, there is no need to use genetically modified organisms, but at what point do conditions favor one or the other? This “tipping point” of this decision has been reached. Indeed, we recognize this point is different for different groups.
Optimization
The ideal scenario is one in which the genetically modified organism’s pathway eliminates toxic intermediates and provides more rapid ultimate degradation (i.e. to CO2 and water), and is completely contained within the project area during and after remediation (spatial and temporal). The rates and products can vary between the best and worst cases, so the selection of the best process requires that all the variables be optimized.
Engineering solutions always involve a combination of actions. For example, in addition to biostimulation with inoculants of microbial strains, the physical conditions of the targeted cleanup site will be changed. Surfactants may be added to facilitate the desorption of otherwise, low aqueous-soluble contaminants (e.g. chlorinated and large molecules) from the solid matrices in soil and sediment.
The soil provides absorbents for the retention of organic compounds, with most sorption surfaces in clay and organic matter. Cation exchange capacity, charge, and the nature of cations on the exchange complexes will determine the amount of sorption of organic compounds. Double layer phenomena often control sorption at the soil particle solid-liquid interface. These thin films around soil particles have thicknesses of 1-10 nm and are composed of ions of opposite charge to the particle. The hydrolysis can be acid or base-catalyzed, depending upon the pH of the soil water and other soil conditions. For example, by adding titanium dioxide particles or dissolved natural organic matter particles to a solution, surface catalyzed, neutral hydrolysis can dominate across a large pH range, even when acid or base catalysis had been observed. Such a relationship is a determinant in controlling remediation.
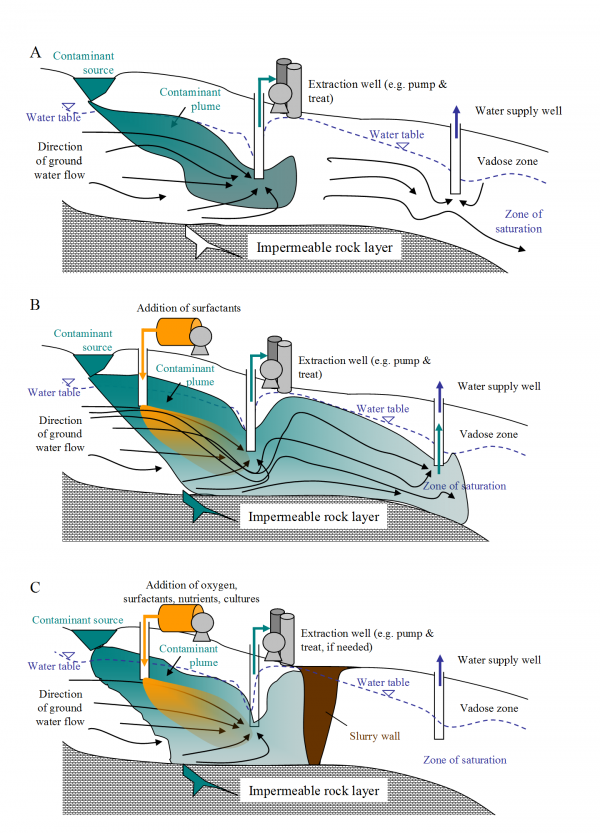
Fig. 6. Changes in contaminant plume by engineering actions. A. Present situation: the plume is moving advectively with flow lines toward the extraction well, but is not yet influenced by the down gradient drinking water well. B. Proposed Action 1: The plume will become more dispersed as the contaminant becomes desorbed from the clay and organic matter in soil by the addition of surfactants; if this action is taken alone, the pollutant will be in the cone of depression of the drinking water well (i.e. contaminating the water supply). C. Proposed Action 2: Systematic control strategy takes advantage of the desorption and entry of the contaminant into the aqueous phase with a slurry wall or other system used to capture and degrade the pollutant in situ adding cultures of a genetically modified bacteria in combination with pump and treat and excavation to ex situ treatment. In addition, the barrier protects the drinking water supply.
The value-added of surfactants is the extent to which they increase the ease at which otherwise nonpolar substances can reach the aqueous phase and come into contact with the biofilms of microbes. This is determined by the structure of the chemical compounds and the concentration as a multiple of critical micellar capacity (CMC). Another systematic consideration is that even though synthetic surfactants can be quite efficient in desorbing recalcitrant compounds, they or the structures they form can be toxic to microorganisms at the concentrations needed for desorption.[14]
Thinking systematically, however, this could present a considerable problem in terms of transport (See Fig.6 A). In other words, since the contaminants are now more dispersed throughout the ground water and soil (Fig. 6 B), have we not made them more mobile and weakened containment? Again, the solution must be systematic. With more of the contaminant dispersed, additional engineering measures must be deployed, such as slurry walls and pumps (See Fig. 6 C.). The resulting treatment will be much better, so long as all of the systems are operating as designed.
These linkages can also be leveraged to provide better results than if each action were taken separately. The key is to apply reliable information to fit the constraints, needs and opportunities of each system.
Footnotes
[1] E. Adler (1997). Lignin chemistry-Past, Present and Future, Wood Science Technology. 11: 169-218.
[2] T. Cajthaml, M. Möder, P. Kacer, V. Šašek and P. Popp (2002). Study of fungal degradation products of polycyclic aromatic hydrocarbons using gas chromatography with ion trap mass spectrometry detection. Journal of Chromatography A. 974: 213-22; M. Mansur, E. Arias, J.L. Copa-Patino, M. Flärdh and A. E. González (2003). The white-rot fungus Pleurotus ostreatus secretes laccase isozymes with different substrate specificities. Mycologia. 95 (6): 1013-20; S.B. Pointing (20010. Feasibility of bioremediation by white-rot fungi. Applied Microbiology Biotechnology. 57: 20-33; and, E. Veignie, C. Rafin, P. Woisel and F. Cazier (2004). Preliminary evidence of the role of hydrogen peroxide in the degradation of benzo[a]pyrene by a non-white rot fungus Fusarium solani. Environmental Pollution 129: 1-4.
[3] See, for example: R. Manimekalai and T. Swaminathan (1999). Optimization of lignin peroxidase production from Phanerochaete chrysosporium using response surface methodology. Bioprocess Engineering. 21: 465-468.
[4] See, for example: O.V. Koroleva, E.V. Stepanova, V.P. Gavrilova, N.S. Yakovleva, E.O. Landesman, I.S. Yavmetdinov and A.I. Yaropolov (2002). Laccase and Mn-peroxidase production by Coriolus hirsutus strain 075 in ajar fermenter. Journal of Biosciences and Bioengineering. 93(5): 449-455.
[5] See, for example: J. Hess, C. Leitner, C. Galhaup, K.D. Kulbe, B. Hinterstoisser, M. Steinwender and Haltrich (2002). Enhanced formation of extracellular laccase activity by the white-rot fungus Trametes multicolor. Applied Biochemistry and Biotechnology. 98-100: 229-241; N. Hatvani and I. Mecs (2001). Production of laccase and manganese peroxidase by Lentinus edodes on malt-containing by-product or the brewing process. Process Biochemistry. 37: 491-496; and I. Herpoel, S. Moukha,, L. Lesage-Meessen, J-C. Sigoillot and M. Asther (2000). Selection of Pycnoporus cinnabarinus strains for laccase production. FEMS Microbiological Letters. 183: 301-306.
[6] Intracellular transport and localization of microsomal cytochrome P450. Analytical & Bioanalytical Chemistry. 392 (6): 1075-1084.
[7] L.P. Wackett (1996). Co-metabolism: is the emperor wearing any clothes? Current Opinion in Biotechnology.7(3); 321-325.
[8] Agency for Toxic Substances and Disease Registry (1996). U.S. Department of Health and Human Services. Toxicological Profile for 2,4,6 Trinitrotoluene. Atlanta, Georgia.
[9] Ibid.
[10] J.C. Spain (1995). Biodegradation of nitroaromatic compounds. Annual Review of Microbiology. 49: 523-555.
[11] Agency for Toxic Substances and Disease Registry (1996); H-Y. Kim and H.G. Song (2003). Transformation and mineralization of 2,4,6-trinitrotoluene by the white rot fungus Irpex lacteus. Applied Microbiology and Biotechnology. 61 (2): 151-156; and, A. M. Ziganshin, A. V. Naumov, E. S. Suvorova, E. A. Naumenko and R. P. Naumova (2007). Hydride-mediated reduction of 2,4,6-trinitrotoluene by yeasts as the way to its deep degradation. Microbiology. 76 (76). 676-682;
[12] S. McFarlan and G. Yao (2009). Anaerobic trinitrotoluene pathway map. University of Minnesota Biocatalysis/Biodegradation Database.
[13] P. Hwang, T. Chow and N.R. Adrian (1998). Transformation of TNT to triaminotoluene by mixed cultures incubated under methanogenic conditions. U.S. Army Corps of Engineers. USACERL Technical Report No. 98/116.
[14] I.V. Robles-González, F. Fava and H.M. Poggi-Varaldo (2008). Review on slurry bioreactors for bioremediation of soils and sediments. Microbial Cell Factories. 7(5). Online.